Hookworm microbiome study: Impact of Experimental Hookworm Infection on the Human Gut Microbiota
Cinzia Cantacessi, Paul Giacomin, John Croese, Martha Zakrzewski, Javier Sotillo, Leisa McCann, Matthew J Nolan, Makedonka Mitreva, Lutz Krause and Alex Loukas. (2014) Impact of Experimental Hookworm Infection on the Human Gut Microbiota. J Infect Dis 3.
Project Summary
The interactions between gastrointestinal parasitic helminths and commensal bacteria are likely to play a pivotal role in the establishment of host-parasite cross-talk, ultimately shaping the development of the intestinal immune system. However, little information is available on the impact of infections by gastrointestinal helminths on the bacterial communities inhabiting the human gut. We used 16S rRNA gene amplification and pyrosequencing to characterize, for the first time to our knowledge, the differences in composition and relative abundance of fecal microbial communities in human subjects prior to and following experimental infection with the blood-feeding intestinal hookworm, Necator americanus. Our data show that, although hookworm infection leads to a minor increase in microbial species richness, no detectable effect is observed on community structure, diversity or relative abundance of individual bacterial species.
Resources
Full manuscript is available at:
Impact of Experimental Hookworm Infection on the Human Gut Microbiota
Cinzia Cantacessi,Paul Giacomin,John Croese,Martha Zakrzewski,Javier Sotillo,Leisa McCann,Matthew J Nolan,Makedonka Mitreva,Lutz Krause, and Alex Loukas. (2014) J. Infect Dis. doi: 10.1093/infdis/jiu256
Raw 16S pyrosequencing data for 8 individuals experimentally infected with N. americanus and tested at two timepoints are available from the NCBI Sequence Read Archive under study accession number SRP041283.
Supplementary table 1, containing relative bacterial abundances per sample, can be downloaded here:
hookworm_microbiome_study-supporting_table_1.xls
Selected Results from the Study
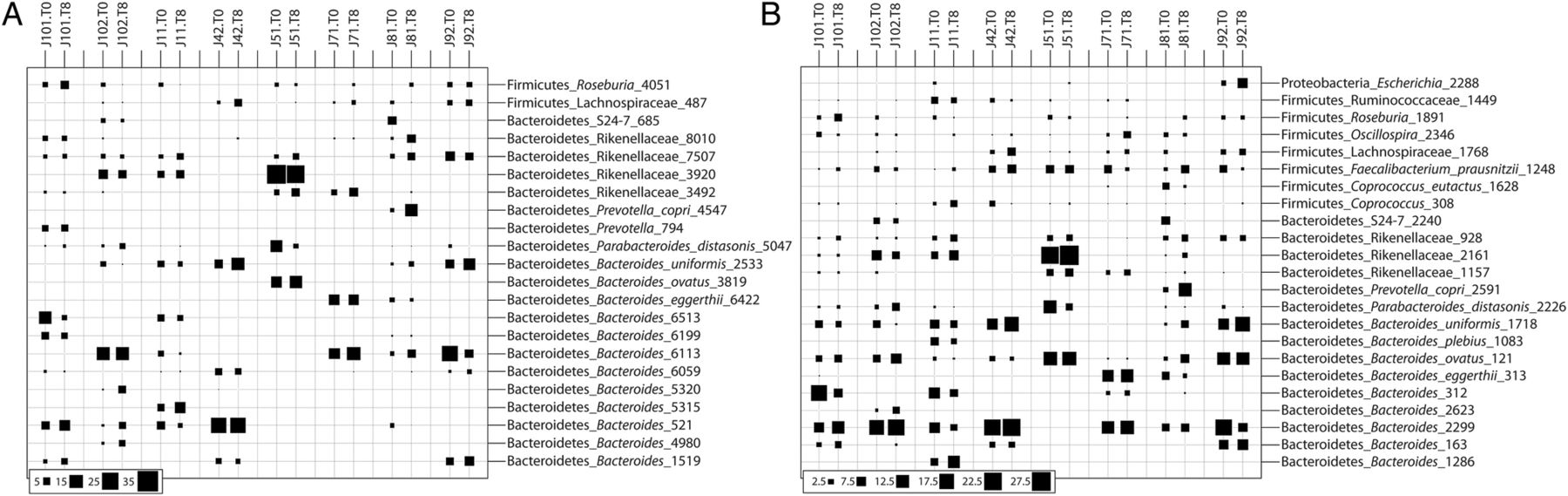
Figure 1. Composition of the fecal microbial communities in patients prior to and following infection by Necator americanus as predicted in the analysis of the V1-V3 (A) and V3-V5 (B) 16S rRNA gene. Bubble sizes reveal the relative abundance (%) of phylotypes (based on 97% sequence identity) in each sample. Codes represent individual subjects included in the study prior to (T0) and at week 8 (T8) post-infection.
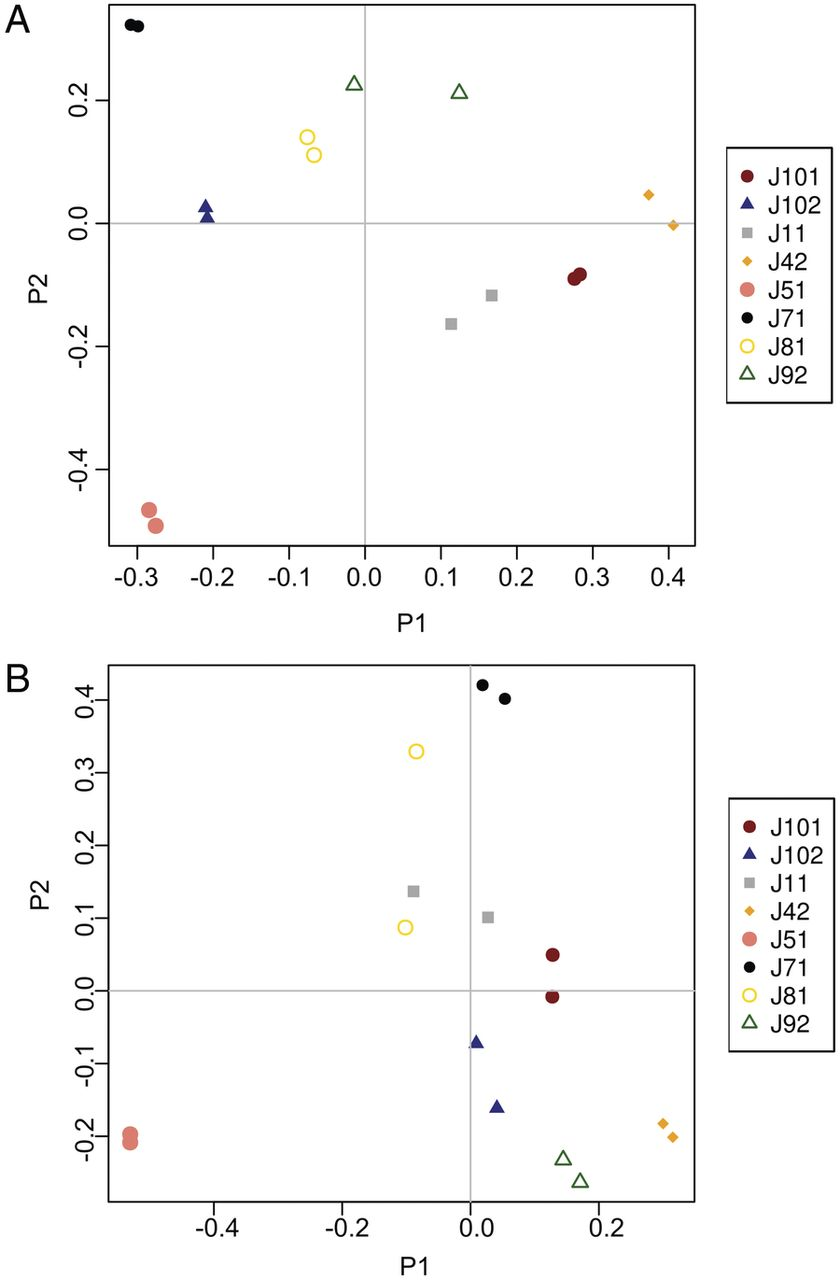
Figure 2. Principal coordinates analysis (PCoA) of the fecal microbiota of human subjects prior to and following experimental infection with Necator americanus, based on analyses of both V1-V3 (A) and V3-V5 (B) hypervariable regions of the prokaryotic 16S rRNA gene. Community similarity was calculated using the Jaccard distance measure of the phylotypes. Codes represent individual subjects included in the study prior to (T0) and at week 8 (T8) post-infection.




